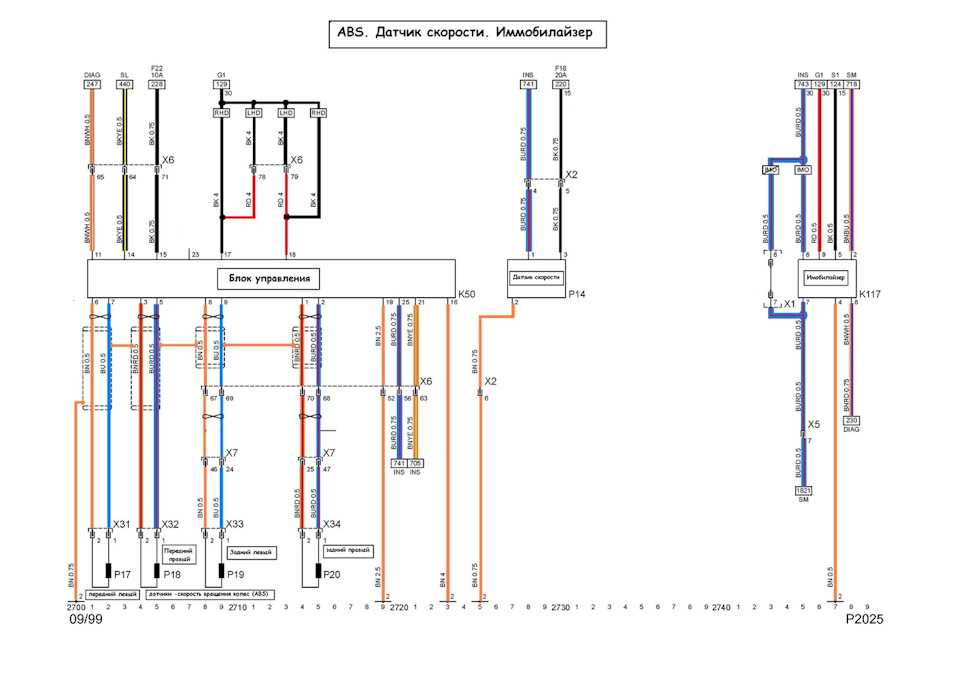
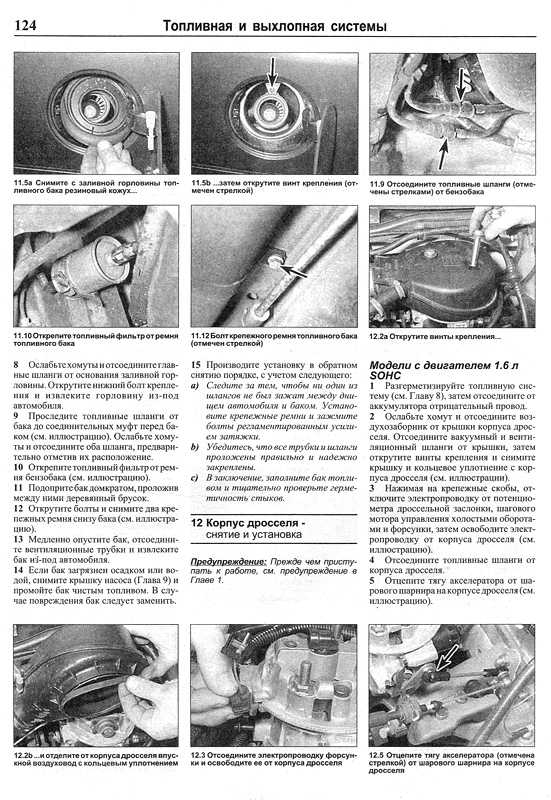
Собрание руководств по ремонту и обслуживанию всех моделей Опель своими руками. Статьи, фото и видео инструкции по ремонту двигателя, кузова и систем авто.

Все про Opel
Обзоры, особенности моделей Opel, ремонт своими руками, тест-драйвы, советы по расходникам и обслуживанию